Prof. Dr. Stephan Urban
Stephan.Urban
@med.uni-heidelberg.de
Phone: +49 6221 56–4902
Fax: +49 6221 56–1946
Development of Envelope Protein-Derived Entry Inhibitors for the Treatment of Acute and Chronic HBV and HDV Infection
Projects
My research interest is molecular mechanisms of Hepatitis B- and Hepatitis D Virus/host interactions with a focus on the early events of infection, identification of hepadnaviral receptors and structural analyses of virus receptor interactions, Development of novel cell culture systems and animal models for HBV/HDV, Clinical development of entry inhibitors (Myrcludex B) for HBV/HDV infection, Development of hepatotropic drugs for the therapy of liver diseases.
1 | Integration of Hepatitis B Virus (HBV) DNA: Underlying mechanisms, its role in pathogenesis, and its uses as a clinical marker
Integration of Hepatitis B Virus (HBV) DNA into the host genome occurs in all known members of the hepadnaviridae family, despite this form not being necessary for the formation of new virions. How HBV DNA integration occurs and its function is completely unknown. Of clinical relevance, HBV DNA integration is associated liver cancer, with the majority of HBV-associated hepatocellular carcinoma (HCC) containing one or more HBV DNA integrations. This is a dramatically higher frequency compared to the 1 intergration per ~103–104 cells in the general hepatocyte population.
In recently published work, we have developed and characterised an in vitro system to reliably detect HBV DNA integrations resultant from a bona fide HBV infection. Using this system and other developed techniques, we intend to determine: i) how HBV DNA integrates; ii) what the consequences of this integration are (particularly in hepatocarcinogenesis); and iii) if these integration events generate detectable biomarkers that can predict liver cancer risk.
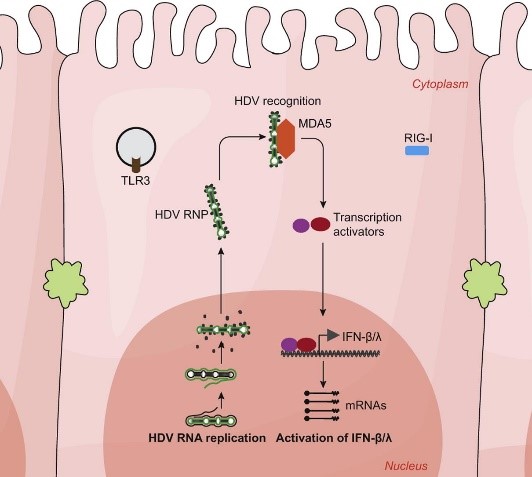
2 | Investigation of the mechanism of innate immunity activation by HDV replication and the role of IFN response in HDV chronicity
In contrast to HBV, HDV infection induces profound innate immune response which is mediated by pattern recognition receptor MDA5 (Zhang, et al. 2018. J Hepatol. 69:25–35). MDA5 is a cytosol sensor usually recognizing long double strand RNA (dsRNA). However, unlike other RNA viruses, HDV replicates its RNA in the nucleus via a unique double-rolling-cycle mechanism without producing long dsRNA. Investigating the innate immune activation during HDV replication will provide important insights for understanding MDA5 mediated innate immune sensing of viral RNA. To this aim, we plan to: (i) identify the HDV RNA ligand recognized by MDA5; (ii) determine the subcellular location of MDA5-HDV RNA interaction; and (iii) elucidate the roles of host factors (LGP2, ADAR1, etc.) and viral factors (HDAg and HBV envelope proteins) in the process of innate immunity activation.
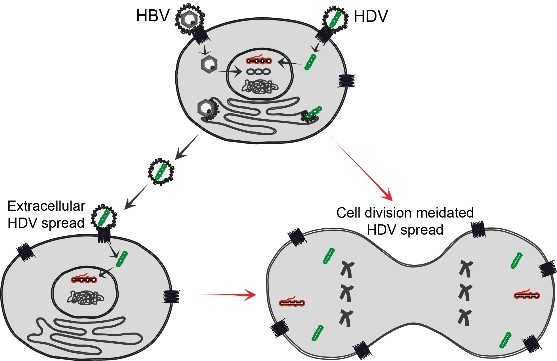
Long term persistence of HBV and HDV makes it challenging to develop curative therapies. Extracellular spreading pathways are important for persistence of both viruses and can be blocked by the entry inhibitor Myrcludex B. However, HDV can also propagate through cell division. Our work has proved that this cell division mediated HDV spread is very sensitive to IFN response (unpublished). In this project we aim to elucidate the mechanism of cell division mediated HDV spread with the focus of understanding the role of IFN response in this process. To this end, we intend to: (i) characterize cell division mediated HDV spread in different cell lines with or without IFN response; (ii) identify the IFN induced antiviral factors contributing the blockade of HDV spread during cell division; (iii) establish an in vitro model supporting both extracellular and cell division mediated HDV spread; and (iv) develop novel synergistic co-treatments by combing inhibitors targeting targeting extracellular HDV spread (MyrB, Lonafarnib, etc.) and cell division mediated HDV spread (IFN, TLR agonists, etc.).
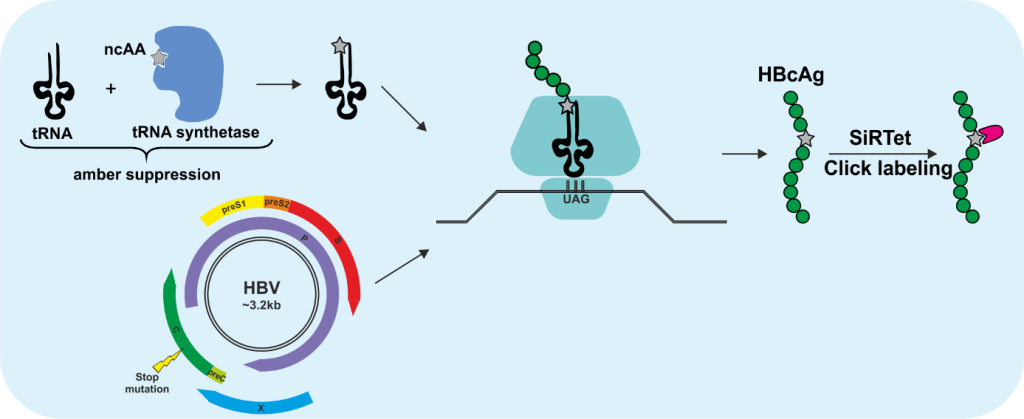
3 | Click labeling of HBV/HDV and the entry receptor NTCP
The incorporation of fluorescent proteins or smaller peptide tags into HBV proteins is very challenging due to the small genome size, the overlapping genes and the tight interactions between not only the capsid monomers but also between capsid and envelope proteins, which is required for the assembly of the virions. Genetic code expansion in combination with click chemistry is an elegant labeling method, especially suited for viruses due to above-mentioned reasons. This technology utilizes the pyrrolysine tRNA/aminoacyl-tRNA synthetase (RS) pair from Methanosarcina mazei, which is orthogonal to the eukaryotic translation machinery and was engineered to enable the incorporation of non-canonical amino acids (ncAA) at the amber stop codon UAG. By utilizing this technology, we introduced ncAA into the core protein of HBV, the delta antigen of HDV and the entry receptor sodium taurocholate cotransporting polypeptide (NTCP), which are subsequently click labeled with functionalized fluorophores. Since the click labeling we utilize is copper-independent, it is also compatible with live-cell imaging. In this project, we aim to create fluorescent tools to visualize not only the virions but also their entry receptor to study (i) trafficking of NTCP with or without the fluorescently labeled preS1 peptide MyrB, (ii) early entry events of HBV and HDV, (iii) intracellular transport of HBV nucleocapsids (HBV-NC) and HDV ribonucleoprotein complexes (HDV-RNP) on their way to the nucleus, and (iv) intracellular trafficking of de novo formed HBV-NC and HDV-RNP.