
Dr. Petr Chlanda
Petr.Chlanda@bioquant.uni-heidelberg.de
BioQuant (INF 267)
Phone: ++49-(0)6221–54-51231
Fax: ++49-(0)6221–54-51480
Membrane Biology of Viral Infection
Projects
We are interested in studying how viruses interact with cellular membranes and lipids during infection. Many enveloped viruses such as influenza or Ebola must cross the cell membrane during entry and exit. To do so, viruses developed or hijacked fascinating protein machinery, which are able to remodel, fuse or cut membranes in processes dependent on specialized lipids. We use and further develop cryo-electron microscopy (cryo-EM) techniques in conjunction with other imaging methods as fluorescence microscopy and imaging mass spectrometry to solve the puzzle of viral-membrane interactions.
1 | Influenza A virus entry and membrane fusion
Influenza A virus (IAV) is a pleiomorphic, enveloped virus that enters the host cell either by endocytosis or macropinocytosis and fuses with the endosomal membrane in a Hemagglutinin (HA)-mediated process that occurs at low pH. Neither the structure of the HA fusion intermediates nor the spatial orientation of the post-fusion HA with respect to the fusion pore are known. We use cryo-ET and subtomogram averaging to further characterize the structure of HA fusion intermediates. In addition, we study viral fusion and disassembly in the cells close to their native state using cryo-CLEM, cryo-FIB/SEM and cryo-ET. This will allow to further investigate the role of host factors such as aggresome machinery, cytoskeleton, IFITMs proteins and lipids such as cholesterol on the membrane fusion and virion disassembly.
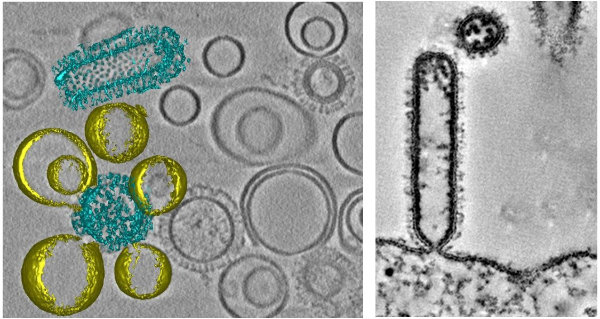
Figure 1 | Membrane fusion of influenza virus-like particles (VLPs) with liposomes (left). Influenza virus assembles and the plasma membrane of the host cell.
2 | Structual analysis of Ebola virus entry intermediates in vitro and in situ
Ebola virus (EBOV) assembles into long filamentous virions (1–15 μm) at the plasma membrane, which upon release, enter epithelial cells by macropinocytosis. The negative single stranded RNA genome is coiled across a length of ~0.9 μm and protected by the nucleocapsid composed of the nucleoprotein (NP), VP35, VP40, and VP24 proteins. The Ebola fusion glycoprotein (GP) is proteolytically processed in the late endosome by low-pH sensitive cathepsin proteases to a 19 kDa fragment, which binds to Niemann-pick-C1 receptor (NPC1). The 19 kDa fragment bound to NPC1 together with yet unknown factor(s) is able to induce membrane fusion allowing the release of the genome. Both the disassembly of the filamentous virus and the mechanism underlying GP mediated membrane fusion in the endosomes are poorly understood processes and have not been structurally characterized. We use non-infectious EBOV VLPs, which are composed of five major structural proteins (GP, NP, VP40, VP35, and VP24) and are structurally similar to EBOV to study EBOV virus entry intermediates by cryo-CLEM, cryo-FIB/SEM and cryo-ET both in vitro and in living cells.
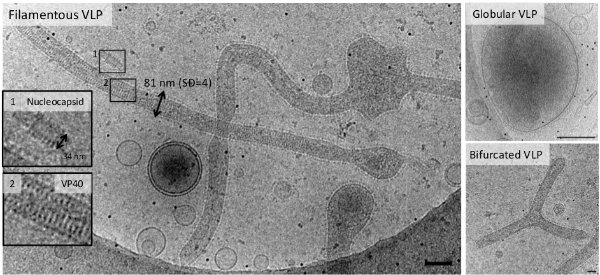
Figure 2 | Ebola virus-like particles are predominantly filamentous.
3 | Spatial lipidomics and viral infection
Secondary ion mass spectrometry (SIMS) allows non-invasive imaging of chemically unmodified lipids with high chemical specificity. However, SIMS has so far been only performed on chemically fixed and dehydrated samples (sample preparation procedures known to severely alter membrane structure). In collaboration with Tom Wirtz (Luxembourg Institute of Science and Technology), we will apply SIMS to vitreous cryo-lamellas of the cells prepared by focused-ion beam milling to provide a spatial map of lipids in cellular organelles at native conditions. The lipid map will be subsequently correlated to the membrane structures observed by cryo-ET. We study lipids such as cholesterol which is crucial in host-pathogen interactions as well as in membrane trafficking.
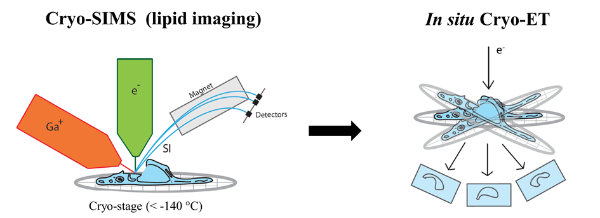
Figure 3 | Correlative cryo-SIMS/ET.
Selected Publications
Winter SL, Chlanda P, (2023) The Art of Viral Membrane Fusion and Penetration, Subcell Biochem, Book: Virus Infected cells. doi: 10.1007/978–3‑031–40086-5_4.
Zimmermann L, Zhao X, Makroczyova J, Wachsmuth-Melm W, Prasad V, Hensel Z, Bartenschlager R, Chlanda P. (2023) SARS-CoV‑2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle. Nature Communications,14(1):7894. doi: 10.1038/s41467-023–43666‑5
Zimmermann L, Chlanda P. Cryo-electron tomography of viral infection — from applications to biosafety. Curr Opin Virol. 2023 Aug;61:101338. doi: 10.1016/j.coviro.2023.101338.
Winter SL, Golani G, Lolicato F, Vallbracht M, Thiyagarajah K, Ahmed SS, Lüchtenborg C, Fackler OT, Brügger B, Hoenen T, Nickel W, Schwarz US, Chlanda P, (2023) The Ebola virus VP40 matrix undergoes endosomal disassembly essential for membrane fusion, EMBO J. 42(11):e113578. doi: 10.15252/embj.2023113578.
Klein S, Golani G, Lolicato F, Beyer D, Herrmann A, Wachsmuth-Melm M, Reddmann N, Brecht R, Lahr C, Hosseinzadeh M, Kolovou A, Schorb M, Schwab Y, Brügger B, Nickel W, Schwarz US, Chlanda P, (2023) IFITM3 blocks viral entry by sorting lipids and stabilizing hemifusion, Cell Host&Microbe, S1931-3128(23)0019–9, doi: 10.1016/j.chom.2023.03.005.
Winter SL, Chlanda P, (2021) Dual-axis Volta phase plate cryo-electron tomography of Ebola virus-like particles reveals actin-VP40 interactions, J Struct Biol, 107742. doi: 10.1016/j.jsb.2021.107742.
Klein S, Wimmer WH, Winter SL, Kolovou A, Laketa V, Chlanda P, (2021) Post-correlation on-lamella cryo-CLEM reveals the membrane architecture of lamellar bodies. Communications Biology, 2021 Jan 29;4(1):137. doi: 10.1038/s42003-020–01567‑z.
Klein S, Wachsmuth-Melm M, Winter SL, Kolovou A, Chlanda P. (2021) Cryo-correlative light and electron microscopy workflow for cryo-focused ion beam milled adherent cells. Methods Mol Biol. Correlative Light and Electron Microscopy, IV Volume 162, Chapter 12.
Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P. (2020) SARS-CoV‑2 structure and replication characterized by in situ cryo-electron tomography. Nature Communications, 11, 5885 doi.org/10.1038/s41467-020–19619‑7.
Klein S, Müller TG, Khalid D, Sonntag-Buck V, Heuser AM, Glass B, Meurer M, Morales I, Schillak A, Freistaedter A, Ambiel I, Winter SL, Zimmermann L, Naumoska T, Bubeck F, Kirrmaier D, Ullrich S, Barreto Miranda I, Anders S, Grimm D, Schnitzler P, Knop M, Kräusslich HG, Dao Thi VL, Börner K, Chlanda P. (2020), SARS-CoV‑2 RNA Extraction Using Magnetic Beads for Rapid Large-Scale Testing by RT-qPCR and RT-LAMP. Viruses. 12(8):863. doi: 10.3390/v12080863.
