Prof. Dr. Mathias Munschauer
mathias.munschauer
(at)med.uni-heidelberg.de
Phone: +49 6221 56–4225
Fax: +49 6221 56–4570
RNA Biology of Viral Infections
Projects
We aim to achieve a highly resolved spatial and temporal understandingof RNA regulatory events that determine the success or failure of viral infection.Particularly, we are interested in learning how viral RNA molecules interface withthe host cell, and how RNA-binding proteins shape the fate of viral RNA to controlreplication, immune evasion, and infection outcome. To dissect these processes,we apply a systems-level, RNA-centric strategy that integrates quantitative massspectrometry, functional genomics, and single-cell transcriptomics data.
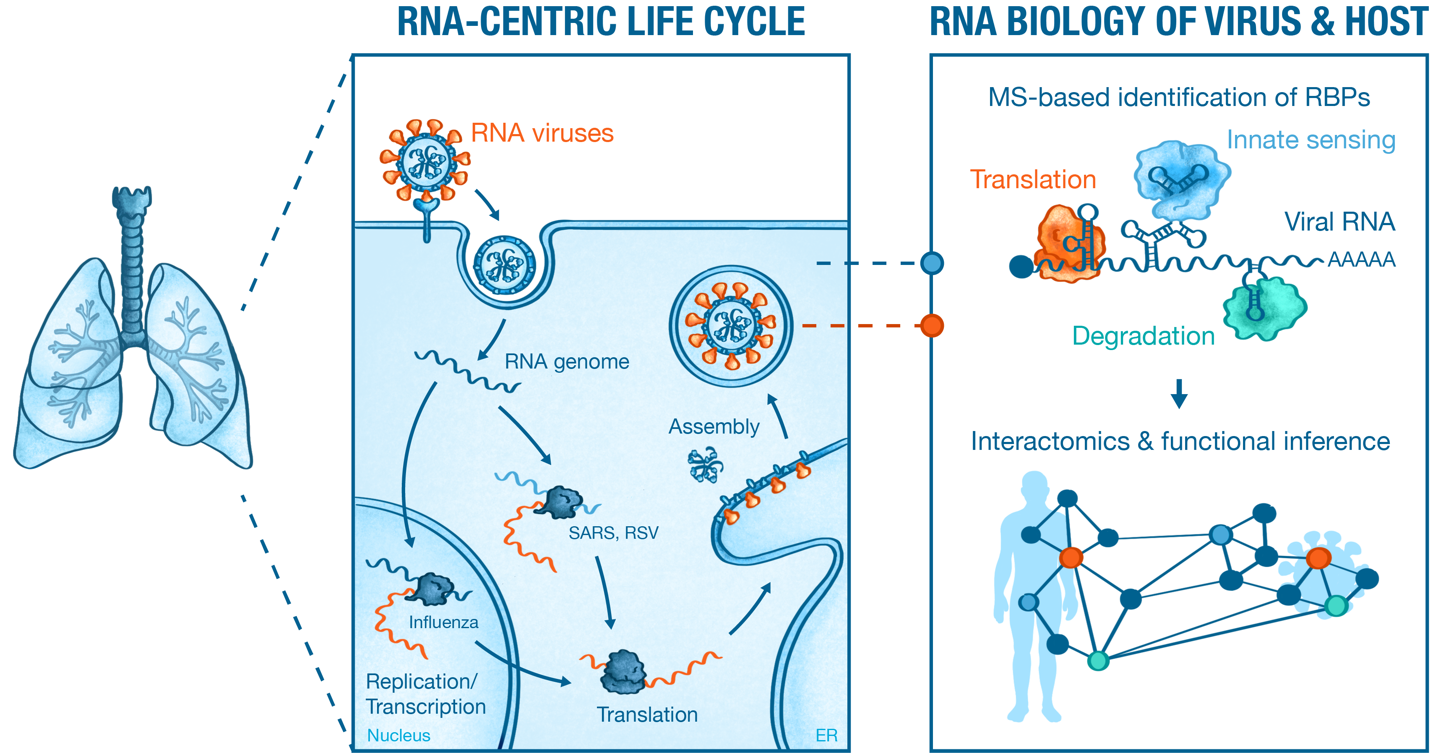
Figure 1. Graphical summary of key research areas in the Munschauer lab.
1 | RNA Interactomics: A New Perspective on RNA Viruses
The systematic analysis of RNA interactomes can uncover molecular signaturesthat detail the functional and regulatory dependencies of specific RNA molecules.Our lab examines these interaction signatures for different RNA viruses at variousstages of the infection cycle to gain insights into the viral RNA replication programand its dependence on specific virus and host cell factors.
We are pioneering a new generation of RNA-centric tools that provideunprecedented insights into the molecular interactions between viruses and theirhosts. One such tool, SHIFTR (https://doi.org/10.1093/nar/gkae038),promises to revolutionize our understanding of viral RNA genomes, their designprinciples, and molecular building blocks. SHIFTR allows us to analyze interactionsfor any cis-regulatory RNA element encoded by any virus, in its native state, andthroughout the viral replication cycle (see Figure 2). In doing so, we are unravelingthe molecular blueprint that dictates the molecular workings of an RNA virus andits reliance on specific interactions the proteomes of virus and host. Current projects in this area aim at developing technologies to assess RNA-proteininteractions and their functional roles at the single-cell and single-molecule level.
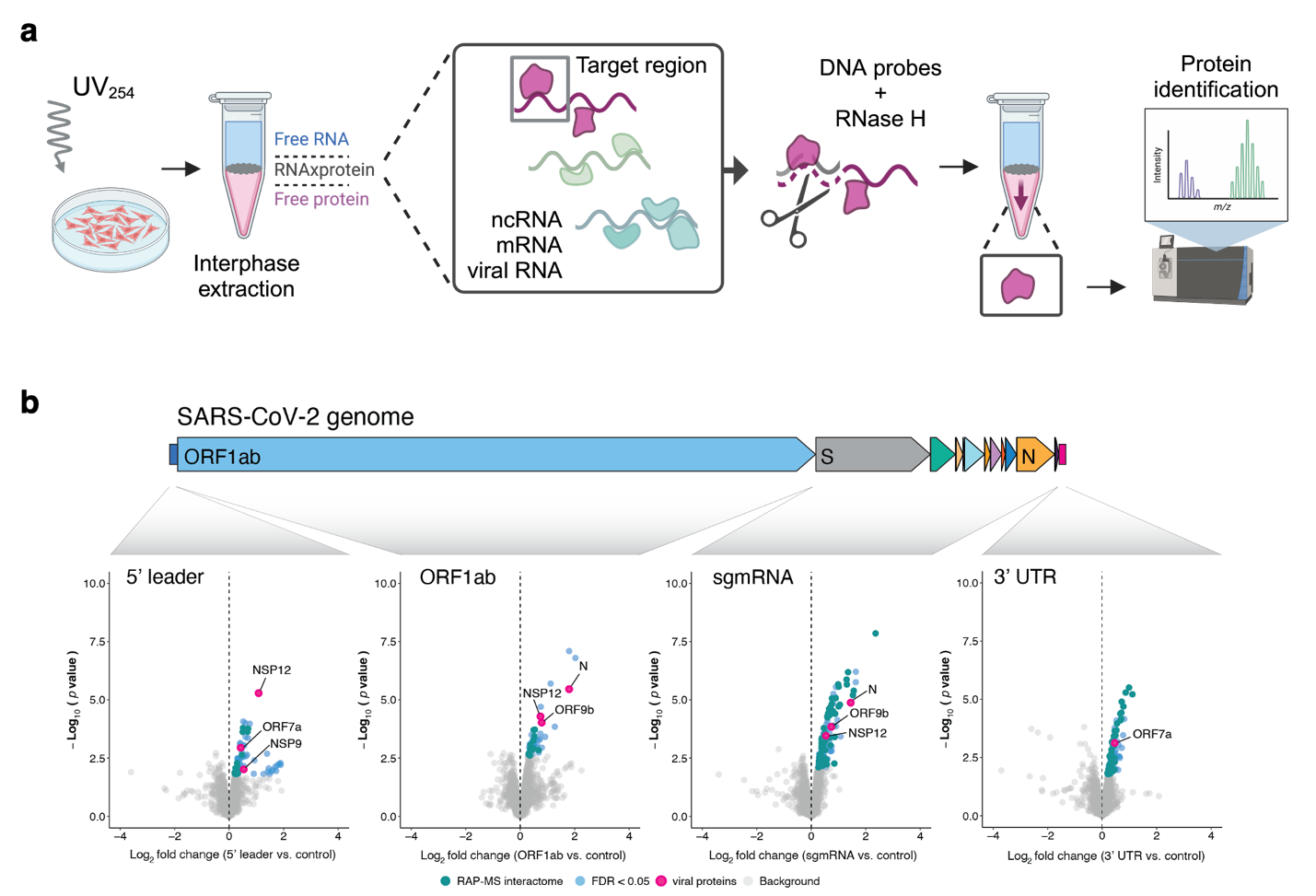
Figure 2. RNA interactomics for defined RNA regions in their native context. a, Graphical outline of the SHIFTR methodology. b, Quantitative mass spectrometrydata for SHIFTR experiments targeting different SARS-CoV‑2 RNA regions.
2 | CRISPR meets Single Cells: From Interaction to Function
Complementing the area of RNA interactomics, we combine CRISPR screeningtechnologies with single-cell transcriptomics (named aka Perturb-seq) tosystematically elucidate the impact of a large number of host factors on the geneexpression programs of viral pathogens and their hosts. Leveraging AI and machinelearning, these single-cell readouts help identify pathways and programs relevantto viral infections and devise strategies to interfere with viral replication. Beyondsingle-cell sequencing, we employ advanced imaging techniques to obtain aspatio-temporally resolved view on the complex molecular biology of viralinfections.
3 | Molecular Mechanisms: a Path to New Antivirals
Our ultimate goal is to characterize the molecular mechanisms underlying (i) thesuccessful execution of viral replication programs in human cells, and (ii) the abilityof a host cell to restrict viral pathogens. Using our unique RNA-centric approach,we profile interactions at unprecedented resolution and resolve functionaldependencies for each viral RNA type, including incoming and replicated viralgenomes, viral mRNAs, as well as viral replication intermediates. This approach hasuncovered fundamental aspects of viral RNA biology, including a previouslyoverlooked protein-priming mechanism utilized by SARS-CoV‑2 (https://doi.org/10.1016/j.cell.2023.09.002), which is unexpectedly controlled by ahost-encoded RNA-binding protein.
4 | RNA Therapeutics
RNA-based therapeutics hold tremendous potential to revolutionize the treatmentof human diseases, including infectious diseases. To improve the design andenhance the biological activity of future RNA-based therapeutics, we investigatehow therapeutic RNAs interact with and are subject to regulation by the RNAregulatory machinery of a target cell. We focus on understanding the sequencefeatures of therapeutic RNAs that determine their stability and activity in the targetcell type, as well as their interaction with the innate immune system.
For additional information on our research projects, please follow the link below:
https://www.klinikum.uni-heidelberg.de/zentrum-fuer-infektiologie/molecular-virology/about-us/research-teams/ag-munschauer

- Aydin J, Gabel A, Zielinski S, Ganskih S, Schmidt N, Hartigan CR, Schenone M,Carr SA, Munschauer M. SHIFTR enables the unbiased identification of proteinsbound to specific RNA regions in live cells. Nucleic Acids Research (2024) gkae038. https://doi.org/10.1093/nar/gkae038
- Schmidt N, Ganskih S, Wei Y, Gabel A, Zielinski S, Keshishian H, Lareau CA,Zimmermann L, Makroczyova J, Pearce C, Krey K, Hennig T, Stegmaier S, Moyon L,Horlacher M, Werner S, Aydin J, Olguin-Nava M, Potabattula R, Kibe A, Dölken L,Smyth RP, Caliskan N, Marsico A, Krempl C, Bodem J, Pichlmair A, Carr SA, ChlandaP, Erhard F, Munschauer M. SND1 binds SARS-CoV‑2 negative-sense RNA andpromotes viral RNA synthesis through NSP9. Cell (2023) 186(22):4834–4850.e23. https://doi.org/10.1016/j.cell.2023.09.002
- Schmidt N, Lareau C, Keshishian H, Ganskih S, Schneider C, Hennig T, Melanson R,Werner S, Wei Y, Zimmer M, Ade J, Kirschner L, Zielinski S, Dölken L, Lander ES,Caliskan N, Fischer U, Vogel J, Carr SA, Bodem J, Munschauer M. The SARS-CoV-2RNA-protein interactome in infected human cells. Nature Microbiology (2021) 6, 339–353. https://doi.org/10.1038/s41564-020–00846‑z
- Munschauer M# (# co-corresponding author), Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, Fulco CP, Subramanian V, Chen J, Ulirch JC,Schenone M, Guttman M, Carr SA, Lander ES#. The NORAD lncRNA assembles atopoisomerase complex critical for genome stability. Nature (2018) 561(7721):132–136. https://doi.org/10.1038/s41586-018‑0453‑z
- Baltz AG*, Munschauer M* (* co-first author), Schwanhaeusser B, Vasile A,Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E,Bonneau R, Selbach M, Dieterich C, and Landthaler M. The mRNA-bound proteome and its global occupancy profile on protein-codingtranscripts. Molecular Cell (2012) 46(5):674–90. https://doi.org/10.1016/j.molcel.2012.05.021
