
Nuclear architecture in viral infection
Projects
We study nuclear architecture and chromatin organization in response to HIV‑1 infection. The nucleus is a complex environment, in which chromatin is organized to support different structural and functional aspects of cell physiology, and represents a challenge for an incoming viral genome, which needs to be integrated into cellular DNA to ensure a productive infection. Integration site selection has functional consequences for viral transcription, which usually follows the integration event. Alternatively, following integration, HIV‑1 can enter the state of latency due to the attenuated or repressed viral transcription. Latency is maintained under current antiviral therapies, but once therapy is interrupted, the virus can resume its productive phase.
1 | HIV‑1 integration and genome organization
a. in depth analysis of the genomic properties of insertion sites in HIV‑1 target cells
One of the main interests of the Lusic lab is understanding the chromatin and genomic features of HIV‑1 integration sites. Although HIV‑1 can in theory target any cellular genomic sequences, and despite the fact it has the well-established preference for transcriptionally active regions of the genome, some genomic regions are targeted much more often than the others. By analysing different independent studies of integration sites both in patients, and in HIV‑1 infections in vitro, we introduced a concept of HIV‑1 recurrent integration genes (RIGs), as genes found in at least two or more independent studies (Marini et al., 2015, Lucic et al 2019). These genes most frequently position in the outer shells of CD4+ T cell nucleus, 1 micron underneath the nuclear envelope, where we also mapped HIV‑1 genome by 3D Immuno-DNA fluorescence in situ hybridization (FISH).
The majority of these recurrent integration genes are classified as cell-type specific genes with super-enhancers (SE), due to which they cluster in 3D nuclear space as revealed by Chromosome conformation capture, coupled with deep sequencing (Hi‑C).
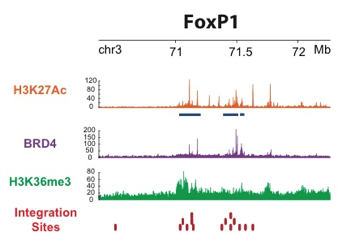
Figure 1 | HIV‑1 integration hot-spots are proximal to super-enhancers. FOXP1 IS (red) superimposition on H3K27ac (orange), SE (blue), H3K36me3 (green) and BRD4 (violet) ChIP-Seq tracks.
b. the reorganization of T cell nucleus upon T cell activation.
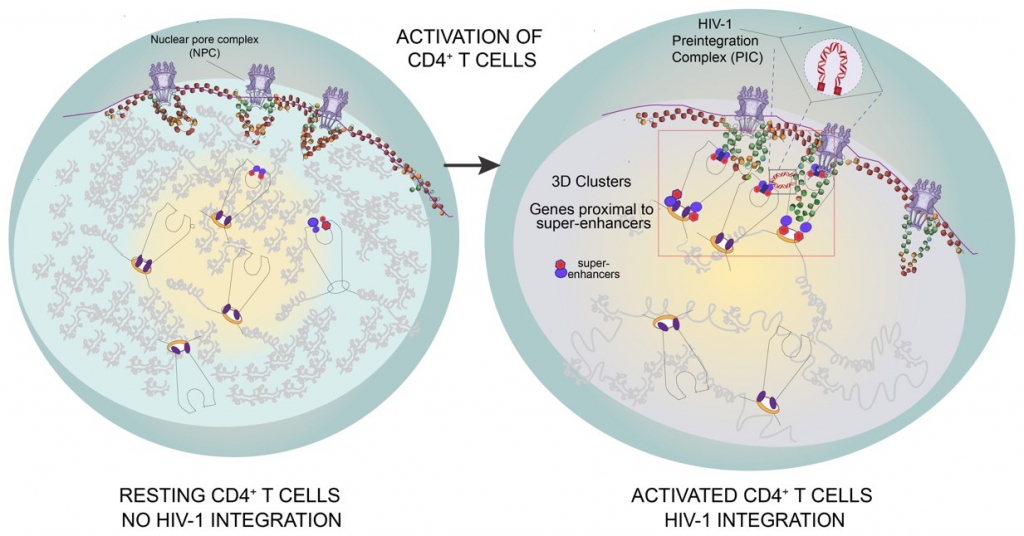
We are particularly interested in how specific genomic elements, such as super-enhancers, control the major transcriptional changes that a resting T cell undergoes when activated. By exploiting transcriptome data of these two primary cell states, and by detecting radial positioning of the cell-type specific genes with distal super-enhancers by 3D Immuno-DNA FISH we could observe a 3D genome reorganization that leads to the docking of these highly expressed SE to the outer shell of the nucleus during activation.

Figure 2 | Genes proximal to super-enhancers change their nuclear positioning upon T cell activation. Three-dimensional immuno-DNA FISH of two RIGs, FOXP1 and STAT5B, in resting and activated (anti-CD3/anti-CD28 beads, IL‑2 for 48 h) CD4+ T cells.
Figure 3 | Model of HIV‑1 integration into the 3D clusters of genes proximal to super-enhancers in activated CD4+ T cells. HIV‑1 integration occurs rarely in resting CD4+ T cells that have large heterochromatin regions and few active genes. HIV‑1 efficiently integrates in activated CD4+ T cells, where 3D clusters of active genes (Recurrent Integration Genes), often proximal to super-enhancers repositioned towards the outer shells of the nucleus, represent insertion hot-spots.
C. Development of a method to capture HIV‑1 integration sites and their chromosomal conformation (DZIF 06.902 and DZIF 04704)
We are establishing a technique that enables the detection of integration sites and contacts with neighboring regions. The first, termed HIV‑1 Cap, has been validated with several historical data sets and histone profiles characteristic for HIV‑1 insertions generated in our laboratory in primary CD4+ T cells (Rheinberger et al., in preparation). We are currently setting up the 4C modification of this method, HIV‑1 GenCap.
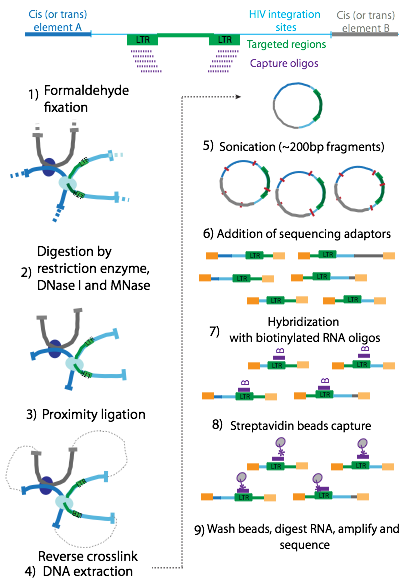
Figure 4 | Schematic representation of HIV Cap and HIV GenCap method. After sonication (5) of the genomic DNA, ends are repaired, adaptors are added and indexes are introduced by PCR (6). The samples are hybridized with biotinylated RNA oligos (7), and captured with streptavidin beads (8 enriched by PCR and sequenced (9). The method can be extended to combine integration site sequencing with a 4C approach (left). For this purpose, DNA is fixated with formaldehyde (1), digested (2) and proximity ligated (3). After reversing the crosslinks, DNA is extracted, sonicated and the Cap method can be performed
d. The role of Nucleoporins (Nups) in nuclear entry, integration site selection and possibly even subsequent transcription of the HIV‑1 genome (Funded by DFG SFB 1129)
The goal of this project is to decipher the role of nuclear pore complexes during HIV‑1 infection and subsequent transcription (more information here). We want to use correlative cryo light and electron microscopy (cCLEM) techniques in collaboration with the Beck and Kräusslich laboratories to visualize the docking of the virus to NPCs (Nuclear Pore Complex) and to determine how exactly it travels through the pore. Once inside, we will use proteomics and genomics approaches to determine how NPC proteins guide integration into RIGs, and how they subsequently define the transcriptional properties of these genes and the integrated HIV‑1 genome in close collaboration with the Beck laboratory.
e. HIV‑1 brain reservoir: how microglia-specific nuclear dynamics affect viral integration and fate (Funded by DFG SPP 2202)
Microglia, brain resident cells of the macrophage lineage, represent a putative HIV‑1 reservoir in the central nervous system. This project aims at discovering HIV‑1 integration and latency patterns in the nuclear environment of the putative reservoir in the brain — microglia. By using the HIV GenCap technique we will define the integration profiles of in vitro infected immortalized primary human microglia cells, isolated from an adult cortical brain surgery (provided by Dr Alvarez-Carbonell). These profiles will be compared to the ones from the closely-related peripheral primary human macrophages. More specifically, we want to understand if lineage-determinant signatures (i.e. PU.1, CTCF) and cell-specific genomics contacts influence viral integration and its fate. These determinants could indeed support immunological inertia of latent cells in the central nervous system and can be addressed to revert latency. This would help to define microglia contribution to HIV‑1 production and persistence under antiretroviral therapy. 3D Immuno FISH (Figure 5) will be used to assess the 3D localization of HIV‑1 as well as the spatial distribution of integration sites; they will be compared to the ones in CD4+ cells.
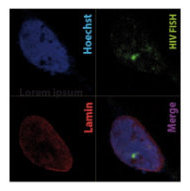
Figure 5 | Microglia model of HIV- 3D Immuno FISH images of integrated HIV (green) in microglia nuclei with confocal microscopy (red lamin, blue Hoechst).
2 | Linking HIV‑1 latency and 3D genome organization
The main obstacle to the functional HIV cure are the latent reservoirs of provirus established in memory CD4+ T cells which cannot be eradicated with current antiviral treatments (cART). Despite some evidence that cART intensification is having positive effects on HIV residual replication, our understanding of HIV‑1 latency and reactivation is still incomplete. While exploring the molecular mechanisms involved in the establishment and maintenance of latency in CD4+ T cells, we obtained substantial evidence that regulation of oxidative phosphorylation pathway and oxidative stress in the cell plays a very important role in HIV‑1 transcriptional control and latency. Our results highlight for the first time the connection and the molecular mechanism between HIV‑1 induced oxidative stress, viral production/latency and Promyelocytic Leukemia Nuclear Bodies (PML NBs) turnover and further strengthen the idea that PML NBs can be used both as molecular markers and/or pharmacologic targets for HIV‑1 infection
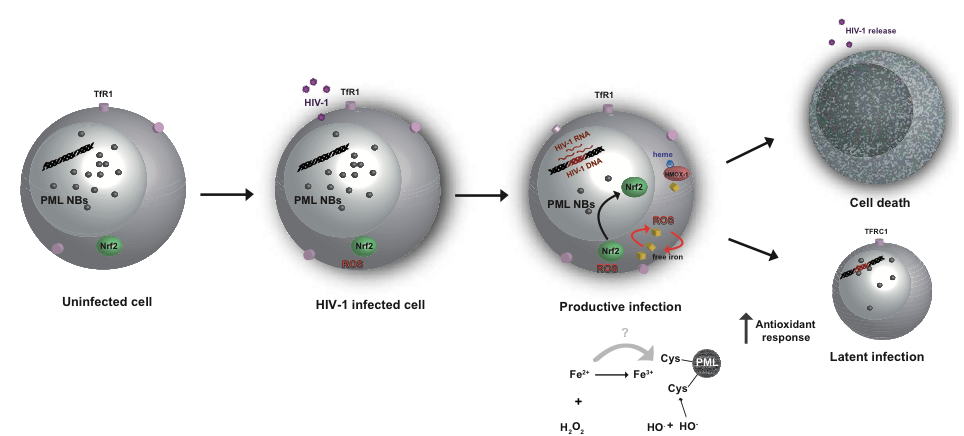
Figure 6 | Regulation of antioxidant responses, intracellular iron content and PML NBs abundance during the transition between productive and latent HIV‑1 infection. In uninfected cells, there is either no or very little oxidative stress, the number of PML Nuclear bodies is normal, and the iron levels are in their steady state. Upon HIV‑1 infection, increased levels of oxidative stress lead to the disbalance in iron homeostasis and heme degradation. In productive infection, there are high levels of oxidative stress, and high antioxidant response, increased iron import followed by decreased PML NB numbers. High antioxidant response ultimately results either in cell death or in iron homeostasis normalization in PML protein levels and PML NB numbers, when latency can be established.
Selected Publications
Complete publication list (PubMed)
Rheinberger M, Costa AL, Kampmann M, Glavas D, Shytaj IL, Sreeram S, Penzo C, Tibroni N, Garcia-Mesa Y, Leskov K, Fackler OT, Vlahovicek K, Karn J, Lucic B, Herrmann C, Lusic M. Genomic profiling of HIV‑1 integration in microglia cells links viral integration to the topologically associated domains. Cell Rep. 2023 Feb 28;42(2):112110. doi: 10.1016/j.celrep.2023.112110. Epub 2023 Feb 13. PMID: 36790927.
Shytaj IL, Fares M, Gallucci L, Lucic B, Tolba MM, Zimmermann L, Adler JM, Xing N, Bushe J, Gruber AD, Ambiel I, Taha Ayoub A, Cortese M, Neufeldt CJ, Stolp B, Sobhy MH, Fathy M, Zhao M, Laketa V, Diaz RS, Sutton RE, Chlanda P, Boulant S, Bartenschlager R, Stanifer ML, Fackler OT, Trimpert J, Savarino A, Lusic M. The FDA-Approved Drug Cobicistat Synergizes with Remdesivir To Inhibit SARS-CoV‑2 Replication In Vitro and Decreases Viral Titers and Disease Progression in Syrian Hamsters. mBio. 2022 Apr 26;13(2):e0370521. doi: 10.1128/mbio.03705–21. Epub 2022 Mar 1. PMID: 35229634; PMCID: PMC8941859.
Zila V, Margiotta E, Turoňová B, Müller TG, Zimmerli CE, Mattei S, Allegretti M, Börner K, Rada J, Müller B, Lusic M, Kräusslich HG, Beck M. Cone-shaped HIV‑1 capsids are transported through intact nuclear pores. Cell. 2021 Feb 18;184(4):1032–1046.e18. doi: 10.1016/j.cell.2021.01.025. Epub 2021 Feb 10. PMID: 33571428; PMCID: PMC7895898.
Lucic B, de Castro IJ, Lusic M. Viruses in the Nucleus. Cold Spring Harb Perspect Biol. 2021 Aug 2;13(8):a039446. doi: 10.1101/cshperspect.a039446. PMID: 33753405; PMCID: PMC8327829.
Lucic B, Wegner J, Stanic M, Jost KL, Lusic M. 3D Immuno-DNA Fluorescence In Situ Hybridization (FISH) for Detection of HIV‑1 and Cellular Genes in Primary CD4+ T Cells. Methods Mol Biol. 2021;2157:239–249. doi: 10.1007/978–1‑0716–0664-3_14. PMID: 32820408.
Fronza, R., Lusic, M., Schmidt, M. and Lucic, B. (2020). Spatial–Temporal Variations in Atmospheric Factors Contribute to SARS-CoV‑2 Outbreak. Viruses 12(6), 588.
Shytaj, I.L., Lucic, B., Forcato, M., Penzo, C., Billingsley, J., Laketa, V., Bosinger, S., Stanic, M., Gregoretti, F., Antonelli, L., Oliva, G., Frese C. K., Trifunovic, A., Galy, B., Eibl, C., Silvestri, G., Bicciato, S., Savarino A. and Lusic M. (2020). Alterations of redox and iron metabolism accompany the development of HIV latency. EMBO J. e102209.
Lucic B, Chen H‑C, Kuzman M, Zorita E„ Wegner J, Minneker V, Wang W, FronzaR, Laufs S, Schmidt M, Stadhouders R, Roukos V, Vlahovicek K, Filion GJ and Lusic M. Spatially clustered loci with multiple enhancers are frequent targets of HIV‑1 Nature Com Accepted for publication. IF 12,353
Muenchau S, Deutsch R, de Castro IJ, Hielscher T, Heber N, Niesler B, Lusic M, Stanifer ML, Boulant S. (2019) Hypoxic environment promotes barrier formation in human intestina epithelial cells through regulation of miRNA-320a expression. Mol Cell Biol. IF 3.813
Ali H, Mano M, Braga L, Naseem A, Marini B, Vu DM, Collesi C, Meroni G, Lusic M, and Giacca, M.(2019) Cellular TRIM33 restrains HIV‑1 infection by targeting viral integrase for proteasomal degradation. Nat Commun, 10:926. IF 12,353
Michieleto D, Lusic M, Orlandini E, Marenduzzo D (2019) Physical principles of retroviral integration. Nat Commun, 10:575. IF 12,353
Bejarano DA, Peng K, Laketa V, Börner K, Jost KL, Lucic B, Glass B, Lusic M, Müller B, Kräusslich HG. (2019) HIV‑1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. Elife. IF 7,616
Lusic M. (2018) Coloring Hidden Viruses. Elife. IF 7,616
Agostini S, Ali H, Vardabasso C, Fittipaldi A, Tasciotti E, Cereseto A, Bugatti A, Rusnati M, Lusic M, Giacca M (2017) Inhibition of Non Canonical HIV‑1 Tat Secretion Through the Cellular Na+,K+-ATPase Blocks HIV‑1 Infection. EBioMedicine. 21:170–181. IF 6.183
Lusic M and Robert F. Siliciano (2017) Nuclear landscape of HIV‑1 infection and integration. Nat Rev Microb. 15:69–82. IF 31,851
Lucic B and Lusic M. (2016) Connecting HIV‑1 integration and transcription: a step toward new treatments. FEBS Letters. 590:1927. IF 2,999
Turrini F, Marelli S, Kajaste-Rudnitski A, Lusic M, Van Lint C, Das AT, Harwig A, Berkhout B, Vicenzi E. (2015) HIV transcriptional silencing caused by TRIM22 inhibition of Sp1 binding to the viral promoter Retrovirology, 12:104. IF 3,867
Marini B, Kertesz-Farkas A, Lucic B, Hashim A, Lisek K, Manganaro L, Pongor S, Luzzati R, Mavilio F, Giacca M and Lusic M (2015) Nuclear architecture dictates HIV‑1 integration site selection Nature, 521: 227–31. IF 41.577
Lusic M and Giacca M (2014) Ground Control to Major Tom: “Prepare for HIV Landing” Cell Host & Microbe, 16: 557–559. IF 17,872
Lusic M and Giacca M (2014) Regulation of HIV‑1 latency by chromatin structure and nuclear architecture. A review article for the special issue “Functional Relevance and Dynamics of Nuclear Organization. J Mol Biol. IF 4.894
Felician G, Collesi C, Lusic M, Martinelli V, Ferro MD, Zentilin L, Zacchigna S, Giacca M (2014) Epigenetic modification at Notch responsive promoters blunts efficacy of inducing notch pathway reactivation after myocardial infarction. Circ Res, 115:636–49. IF 15,211
Lusic M, Marini B, Ali H, Lucic B, Luzzati R, and Giacca M (2013) Proximity to PML Nuclear Bodies negatively regulates HIV‑1 gene expression in CD4+ T cells. Cell Host & Microbe, 13: 665–677. Research highlight in Science Vol 341 (2013) and in Cell Host & Microbe 13:625–626. IF 17,872
Della Chiara G, Crotti A, Liboi E, Giacca M, Poli G and Lusic M. (2011) Negative Regulation of HIV- 1 Transcription by a Heterodimeric NF-kB1/p50 and C‑Terminally Truncated STAT5 Complex. J Mol Biol, 410: 933–943. IF 4,894
Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M and Cereseto A (2011) KAP‑1 inhibits HIV‑1 integration. Cell Host & Microbe, 9: 484–95. IF 17,872
Manganaro L, Lusic M*, Gutierrez MI, Cereseto A, Del Sal G and Giacca M* (2010) Concerted action of cellular JNK and Pin‑1 restricts HIV‑1 genome integration to activated CD4+ T lymphocytes. Nat Medicine, 16: 329–323 (* corresponding authors). IF 32,621
Dieudonné M, Maiuri P, Biancotto C, Knezevich A, Kula A,Lusic M, and Marcello A. (2009) Transcriptional competence of the integrated HIV‑1 provirus at the nuclear periphery EMBO J. 28:2231–2243. IF 9,792
Sabo’ A, Lusic M, Cereseto A and Giacca M (2008) Acetylation of conserved lysines in the catalytic core of CDK9 regulates kinase activity and subnuclear localization. Mol Cell Biol, 28:2201–12 . IF 3,813
Vardabasso C, Manganaro L, Lusic M, Marcello A and Giacca M (2008) The histone chaperone protein Nucleosome Assembly Protein‑1 (hNAP‑1) binds HIV‑1 Tat and promotes viral transcription. Retrovirology, 28:5–8. IF 3,867
Perkins KJ, Lusic M, Mitar I, Giacca M and Proudfoot NJ (2008) Transcription dependent gene looping of the HIV‑1 provirus is dictated by recognition of pre-mRNA processing signals. Molecular Cell, 29:56–68. IF 14,248
