
Dr. Markus Ganter
markus.ganter
(at)med.uni-heidelberg.de
Phone: +49 6221 56–6546
Fax: +49 6221 56–5751
Nuclear Autonomy in Multinucleated Cells
Projects
Cells usually multiply by duplicating their genome and divide themselves into two daughter cells. Yet, in some organisms, cellular multiplication follows distinct, somewhat bizarre routes. One of these rather unusual ways can be seen in Plasmodium spp., the causative agent of malaria. An unusual feature of Plasmodiumreplication inside erythrocytes, known as schizogony, is the number of nuclei and, hence, daughter cells they produce during one round of intracellular replication. Parasites with odd numbers of nuclei can be readily seen (Fig. 1). Divergence from a geometric expansion (e. 2, 4, 8, 16, or 32) is caused by asynchronous replication, suggesting that individual nuclei replicate autonomously until a global mechanism takes over and co-ordinates daughter cell formation. This is in striking contrast with nuclear division in other multinucleated cells, such as the early Drosophila embryo, where nuclei follow a geometric expansion, as their division is relatively synchronous. To gain insight into the regulation of Plasmodium falciparum replication, we conducted a forward genetic screen for the conditional protein expression of protein kinases and identified a kinase as essential for proliferation (Fig. 2). This kinase is Plasmodium-specific and a crucial regulator of the continuous rounds of DNA replication, histone modification, and regulation of gene expression. We also found that this kinase is required for transmission to the mosquito. This work serves as a starting point to gain a better understanding of nuclear autonomy in Plasmodium.

Figure 1 | Nuclei divide autonomously in Plasmodium. Odd numbers of nuclei can be readily seen during the development of P. falciparum.
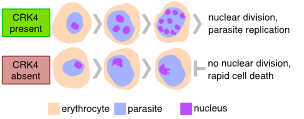
Figure 2 | Plasmodium falciparum CRK4 is essential for replication in the pathogenic blood stage of infection.
1 | Analysis the spatiotemporal parameters of nuclear autonomy
To analyze the spatiotemporal parameters of nuclear autonomy, we are using live-cell microscopy. In a set of reporter parasites, we determine the dynamics of DNA replication, nuclear division, and organelle development. These data inform on a mathematical model, describing nuclear autonomy in P. falciparum (Fig. 3).
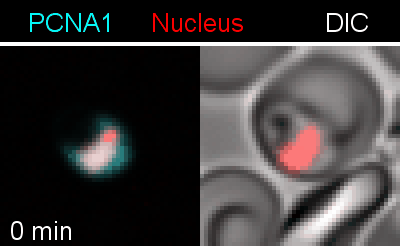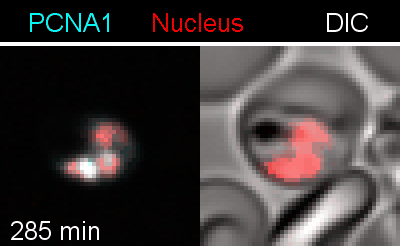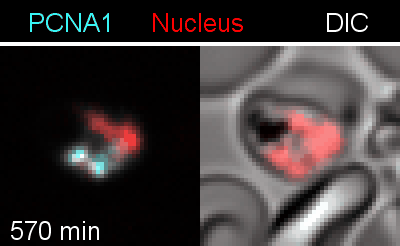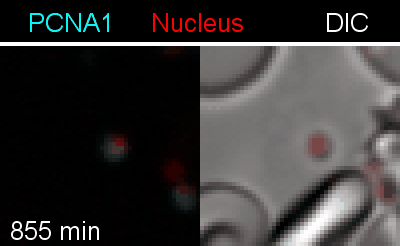
Figure 3 | Plasmodium falciparum reporter cell line. The nuclei of the parasites are marked with a red-fluorescent protein and a component of the DNA replication machinery (PCNA) is marked in cyan.
2 | Regulation of autonomous nuclei replication
To understand how autonomous nuclei regulate their replication, we want to describe the signalling cascade that orchestrates DNA replication in individual nuclei. For this we are using the mayor S‑phase regulator PfCRK4 as a starting point and are currently defining its interactome. This work will inform on substrates of the kinase as well as on upstream regulators.
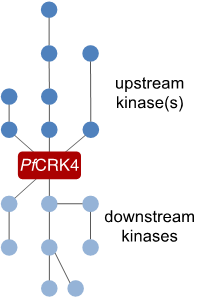
Figure 4 | Cartoon representing the elusive signalling cascade, which triggers PfCRK4.
3 | Function of regulators and effectors in asynchromous DNA replication
Our previous work suggests that a set of proteins of unknown function is involved in DNA replication and nuclear division. These proteins were identified through phosphoproteomic profiling of PfCRK4 and showed a much-reduced phosphorylation in absence of the kinase. To characterize the function of these regulators and effectors involved in asynchronous DNA replication, we are using reverse genetic tools and inducible gene-depletion approaches.
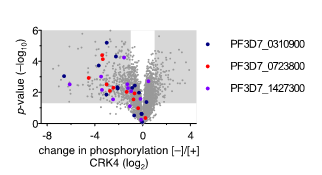
Figure 5 | Change in phosphorylation of three proteins of unknown function in absence of CRK4. Phosphoproteomic profiling of PfCRK4-depleted parasites. Grey areas indicate phosphopeptides with a ≥2‑fold change in phosphorylation and p<0.05.
4 | Collaboration with Zendia GmbH
In collaboration with the biomedical start-up company Zendia GmbH and the Frischknecht lab at CIID, we are developing a novel rapid diagnostic test (RDT) to detect infections with Plasmodium falciparum. Unlike other RDTs, this system is able to enrich and detect parasites from peripheral blood, allowing for an early onset of anti-malaria treatment.
Pape C., Remme R., Wolny A., Olberg S., Wolf S., Cerrone L., Cortese M., Klaus S., Lucic B., Ullrich S., Anders-Össwein M., Wolf S., Berati C., Neufeld C., Ganter M., Schnitzler P., Merle U., Lusic M., Boulant S., Stanifer M., Bartenschlager R., Hamprecht F.A., Kreshuk A., Tischer C., Kräusslich H‑G., Müller B. and Laketa V. Microscopy-based assay for semi-quantitative detection of SARS-CoV‑2 specific antibodies in human sera. BioAssays, 2021; 200025
Quadt K., Smyrnakou X., Frischknecht F., Böse G., Ganter M. Plasmodium falciparum parasites exit the infected erythrocyte after haemolysis with saponin and streptolysin O. Parasitology Research. 119:4297–4302
Przyborski J. and Ganter M. (2018). Das ungewöhnliche Leben der Malariaparasiten. Biol. Unserer Zeit 48, 162–169 (invited review article in German)
Ganter M., Goldberg J.M., Dvorin J.D., Paulo J.A., King J.G., Tripathi A.K., Paul A.S., Yang J., Coppens I., Jiang R.H.Y., Elsworth B., Baker D.A., Dinglasan R.R., Gygi S.P., Duraisingh M.T. (2017). Plasmodium falciparum CRK4 directs continuous rounds of DNA replication during schizogony. Nat. Microbiol. 2(5):17017
Paul A.S., Saha S., Engelberg K., Jiang R.H.Y., Coleman B.I., Kosber A.L., Chen C., Ganter M., Espy N., Gilberger T.W., Gubbels M.J., Duraisingh M.T. (2015). Parasite calcineurin regulates host cell recognition and attachment by apicomplexans. Cell Host Microbe. 18:49–60
Ganter M., Rizopoulos Z., Schuler H., Matuschewski K. (2015). Pivotal and distinct role for Plasmodium actin capping protein alpha during blood infection of the Malaria parasite. Mol. Microbiol. 96:84–94
Coleman B.I., Skillman K.M., Jiang R.H.Y., Childs L.M., Altenhofen L.M., Ganter M., Leung Y., Goldowitz I., Kafsack B.F.C., Marti M., Llinas M., Buckee C.O., Duraisingh M.T. (2014). A Plasmodium falciparum histone deacetylase regulates antigenic variation and gametocyte conversion. Cell Host Microbe. 16:177–186
*Sattler J., *Ganter M., Hliscs M., Matuschewski K., Schuler H. (2011). Actin regulation in the malaria parasite. Eur. J. Cell Biol. 90:966–971 (*equal contribution)
*Siden-Kiamos I., *Ganter M., Kunze A., Hliscs M., Steinbüchel M., Mendoza J., Sinden R., Louis K., Matuschewski K. (2011). Stage-specific depletion of myosin A supports an essential role in motility of malarial ookinetes. Cell Microbiol. 13(12):1996–2006 (*equal contribution)
Ganter M., Schuler H., Matuschweski K. (2009). Vital role for the Plasmodium capping protein (CP) beta subunit in motility of malaria sporozoites. Mol. Microbiol. 74:1356–1367
Kursula I., Kursula P., Ganter M., Panjikar S., Matuschewski K., Schuler H. (2008). Structural basis for parasite-specific functions of the divergent profilin of Plasmodium falciparum. Structure. 16:1638–1648.
